course_model
Principle Component Analysis (PCA)
PCA is a method for reducing the number of columns while retaining as much information as possible.
Outcomes
- Understand the PCA algorithm
- Apply PCA for dimensionality reduction
PCA Algorithm
Principal Component Analysis (PCA) is a technique for:
- Reducing dimensionality (many features → fewer features),
- While preserving as much variance as possible,
- By creating new, uncorrelated features called principal components.
Each principal component is:
- A linear combination of the original features.
- Orthogonal (perpendicular) to the others.
-
Ordered so that:
- PC1 explains the most variance,
- PC2 explains the second most variance,
- etc.
import numpy as np
import pandas as pd
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
# Create a dataset
df = pd.DataFrame({
'height': [10, 12, 13, 15, 18, 20, 22, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70],
'weight': [30, 32, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, 100, 105],
'age': np.random.randint(20, 60, size=17)
})
# PCA works best when features are centered; standardization is often used when units differ.
X = df[['height', 'weight', 'age']].values # shape: (17, 3)
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
# `X_scaled` has mean ~0 and variance ~1 in each column.
# Fit PCA
pca = PCA(n_components=2)
X_pca = pca.fit_transform(X_scaled)
# `pca.components_` is a 2×3 matrix. Each row is a principal component (PC1 and PC2), each column corresponds to a feature (`height`, `weight`, `age`).
# `pca.explained_variance_ratio_` might look something like `[0.95, 0.05]`, meaning the amount of variance captured by each principal component.
print("Principal components (directions):")
print(pca.components_)
print("\nExplained variance ratio:")
print(pca.explained_variance_ratio_)
Principal components (directions):
[[ 0.70317421 0.70333577 -0.10423451]
[ 0.07527239 0.07213746 0.99455028]]
Explained variance ratio:
[0.66635142 0.32967702]
Add PCA results back to a DataFrame
We can now use the PCA results to do further analysis or visualization.
df_pca = pd.DataFrame(
X_pca,
columns=['PC1', 'PC2']
)
df_with_pca = pd.concat([df.reset_index(drop=True), df_pca], axis=1)
import matplotlib.pyplot as plt
# Plot PCA-transformed data with two scatter calls (matplotlib scatter marker must be a single style)
plt.figure(figsize=(12, 5))
young = df_with_pca[df_with_pca['age'] < 30]
middle = df_with_pca[(df_with_pca['age'] >= 30) & (df_with_pca['age'] < 45)]
old = df_with_pca[df_with_pca['age'] >= 45]
plt.scatter(
young['PC1'], young['PC2'],
c=young['height'] + young['weight'],
marker='.', cmap='viridis', alpha=0.7, label='age < 30'
)
plt.scatter(
middle['PC1'], middle['PC2'],
c=middle['height'] + middle['weight'],
marker='*', cmap='viridis', alpha=0.7, label='30 ≤ age < 45'
)
plt.scatter(
old['PC1'], old['PC2'],
c=old['height'] + old['weight'],
marker='o', cmap='viridis', alpha=0.7, label='age ≥ 45'
)
plt.colorbar(label='height + weight')
plt.xlabel('PC1')
plt.ylabel('PC2')
plt.title('PCA Scatter (Age-Based Markers)')
plt.legend()
plt.tight_layout()
plt.show()
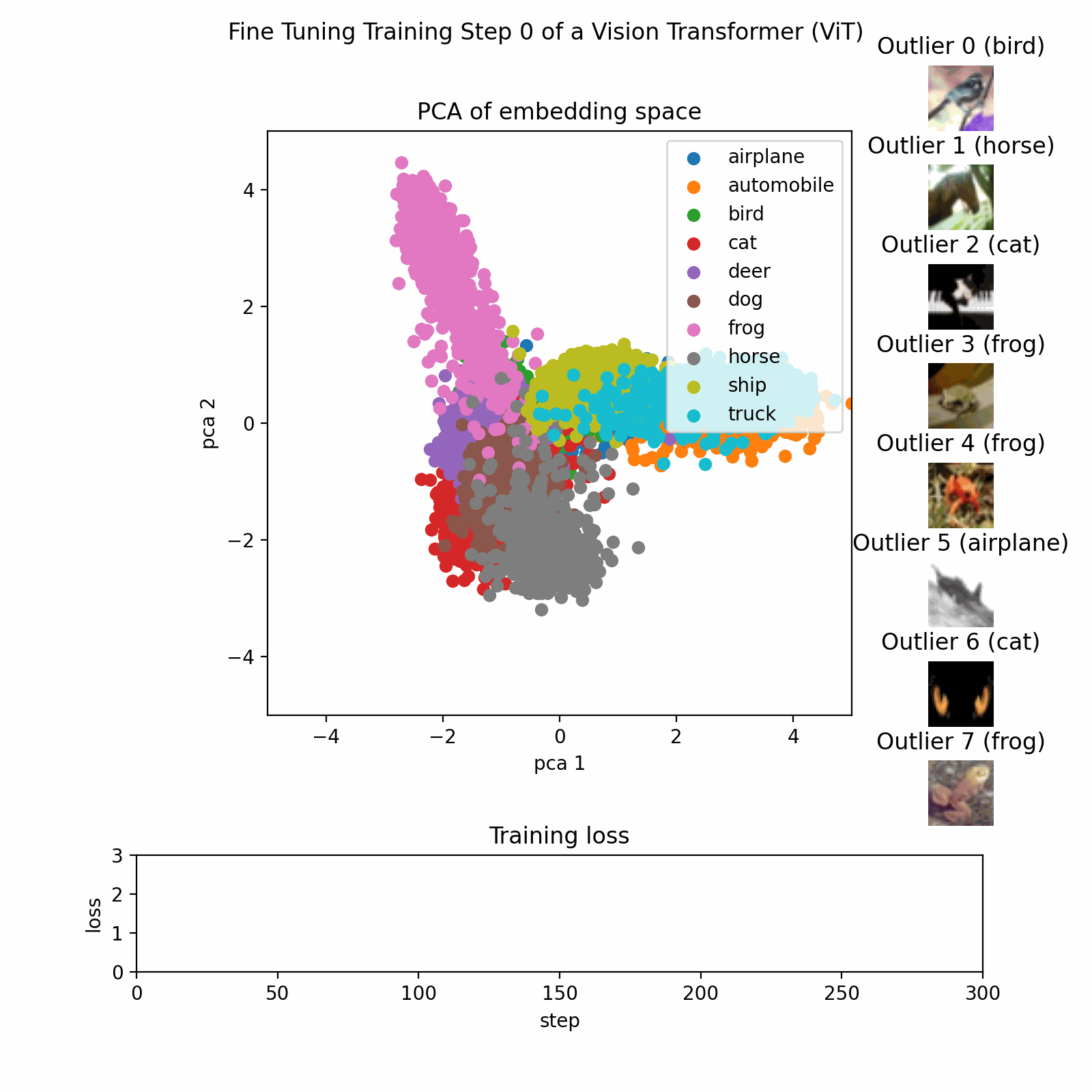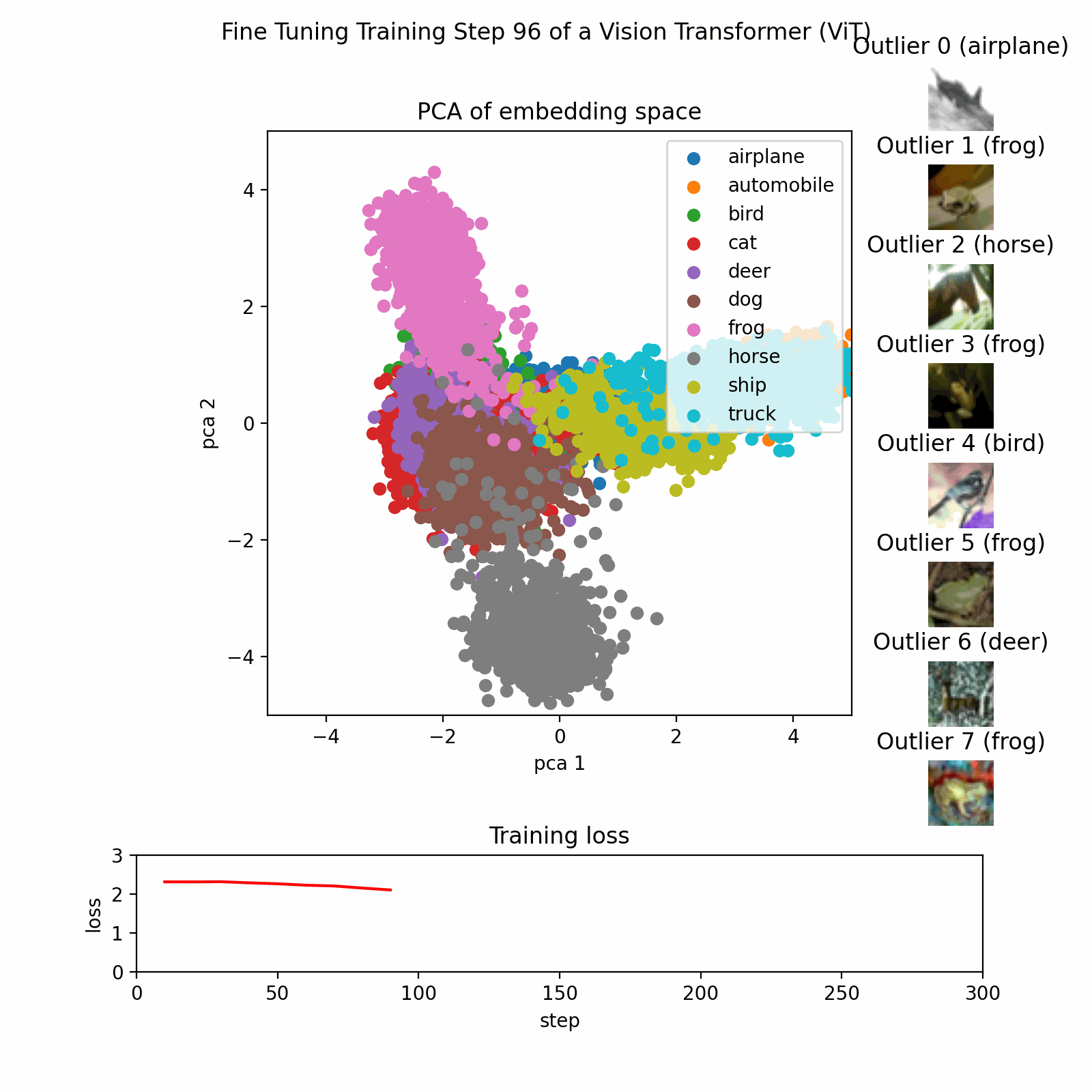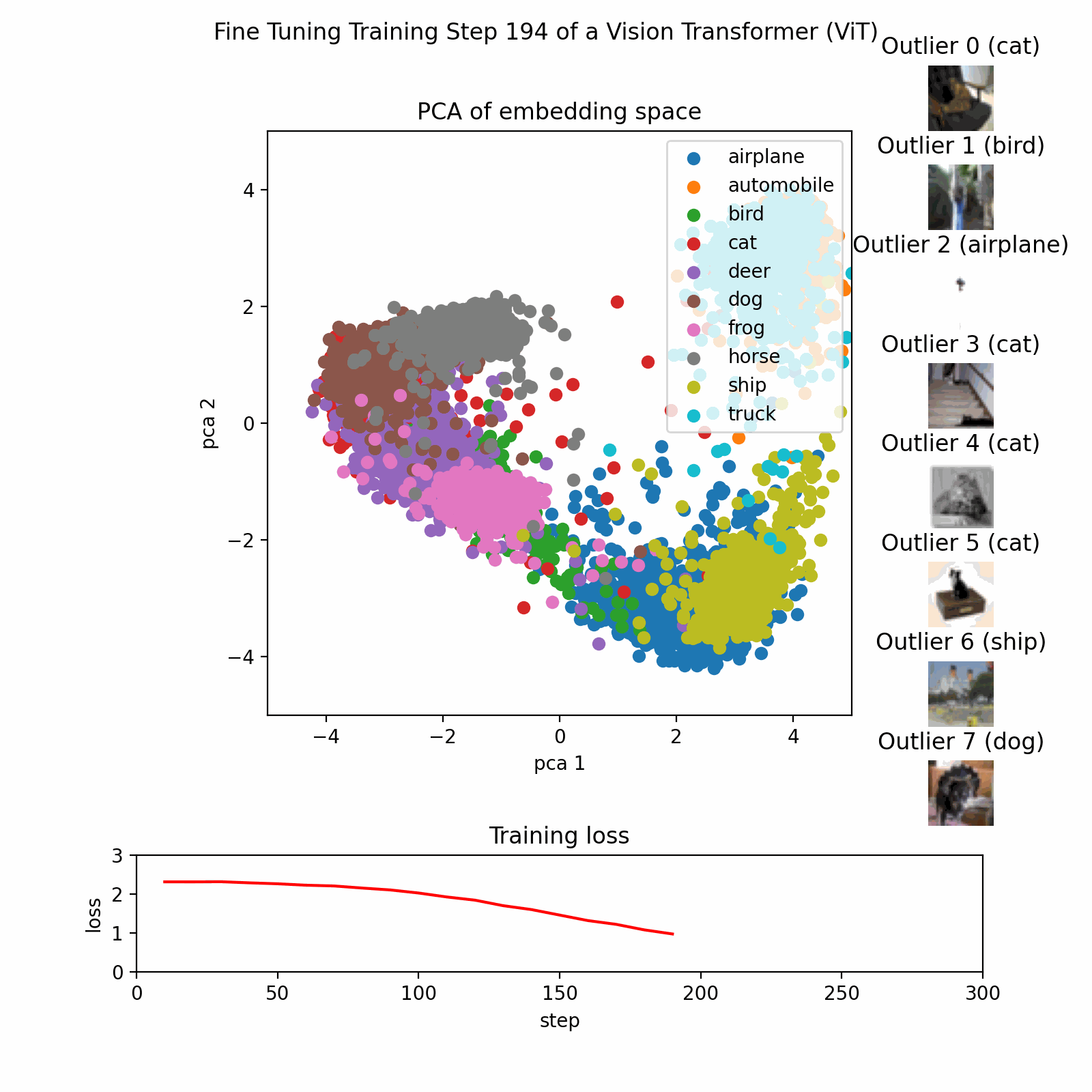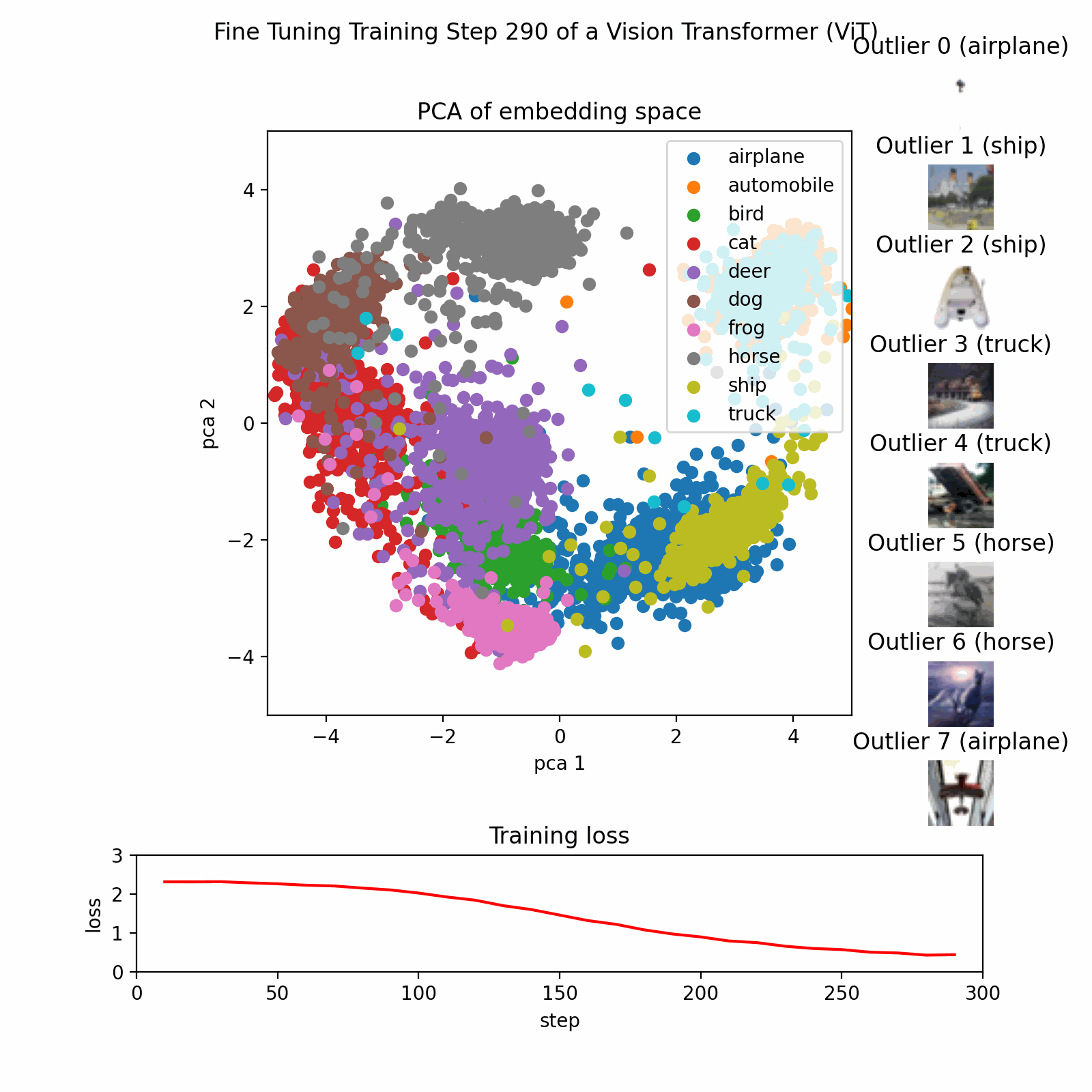
Example PCA Process
This image show the PCA training process on a dataset with three features reduced to two principal components.